Comparative genomics of Citrus-associated Phyllosticta species
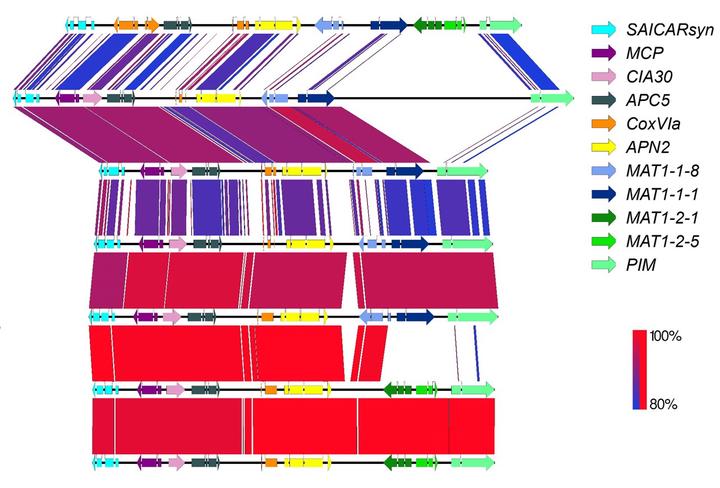 Pairwise comparison of mating-type and flaking genes between Phyllosticta species
Pairwise comparison of mating-type and flaking genes between Phyllosticta species‘Phyllosticta genus comprises several plant pathogenic species that are responsible for diseases symptoms such as leaf and fruit spots in many different hosts, as well as endophytic and saprophytic species. With the recent description of P. paracapitalensis and P. paracitricarpa, eight Phyllosticta are now known to be associated with Citrus hosts, either as pathogens or as endophytes, presenting different interactions and lifestyles. As this kind of variation in host-interaction within so closely related species exist, these Phyllosticta species provide a good model for evolutionary studies that aim to understand the evolution of phytopathogens, as well as obtain new targets for functional studies.
Although significant progress has been achieved in understanding the Phyllosticta genus, many questions still remain open. Even though pathogenic species have been described as causing Citrus Tan Spot, only P. citricarpa is regarded as the causative agent of Citrus Black Spot. These differences in biology and ecology of the citrus-associated Phyllosticta species have not yet been addressed from an evolutionary and genomic perspective. Such kind of approach could be useful in investigating and understanding the biological processes involved in the different associations that these species develop with citrus plants. Moreover, these analyses could help to detect possible species-specific adaptation and shared pathogenicity related traits between the Phyllosticta species, as well as unique features that could explain the pathogenicity of P. citricarpa.
As the genomes of the citrus-associated Phyllosticta species became available (with the exception of P. paracapitalensis), it is now possible to perform comparative analyses to understand functional and evolutionary differences between these species. Thus, the main goal of this project is to perform comparative genomic analyses for the citrus-associated Phyllosticta species, focusing on the evolution of pathogenicity-related mechanisms mainly for P. citricarpa. We are mostly interested in genome evolution and pathogenicity-genes evolution, with an emphasis on the identification of genome-wide patterns and signatures of positive selection between these closely related Phyllosticta species, and how these patterns could be related to the pathogenicity of P. citricarpa. Based in these genome analyses, we intend to further explore this topic through functional approaches involving gene deletion/silencing and transcriptome analysis, in order to validate the results and assess the involvement of positively-selected genes in pathogenicity.
This project is conducted at UFPR under supervision of Prof. Chirlei Glienke and at the MPI for Evolutionary Biology, under supervision of Prof. Eva Stukenbrock, in collaboration with Prof. Marcos Machado from IAC (Instituto Agronômico de Campinas, Brazil), Prof. Pedro Crous and Prof. Ewald Groenewald (Westerdijk Fungal Biodiversity Institute).’
